Understanding the genetic makeup of food crops is critical if we want to develop sustainable ways of protecting those crops against disease. To this end, researchers are using genomics to study plants and their characteristics. Distinguished McKnight Professor Nevin Young (Plant Pathology), together with Associate Professor Peter Tiffin (Plant Biology), Professor Mike Sadowsky (Soil, Water and Climate), and Assistant Professor Bob Stupar (Agronomy and Plant Genetics) have been using MSI for many years as part of their investigations into legumes, the family of plants that includes soybeans, peas, and alfalfa. Legumes are especially interesting in that they form symbiotic relationships with rhizobial bacteria and arbuscular mycorrhizal fungi. These symbiotic relationships allow legumes to extract compounds such as nitrogen and phosphorus out of the soil and provide natural forms of fertilizer for agriculture.
Medicago truncatula (barrel medic) is used by many researchers as a model for legume genomics. The Young group collaborates with research groups worldwide in studying M. truncatula, using massive genomic sequencing methods to study the gene systems and genomic variations that impact these valuable plant-microbe interactions.
MSI’s RISS group is working with the Young lab on this NSF-funded research. Dr. Kevin Silverstein, Operations Manager and Scientific Lead of the RISS group, is the co-PI on the most recent NSF grant to fund this work. The researchers are using MSI’s Itasca system to map the immense collection of raw sequencing data generated by the project and they are also using the Institute’s high-performance storage capabilities. They have also developed graph analytic applications that offer the opportunity to extend MSI’s compute resources in the area of bioinformatics.
The Young group has published numerous papers about their research. A sampling of recent papers includes:
- “Phylogenetic signal variation in the genomes of the genus Medicago (Fabaceae),” JB Yoder, R Briskine, J Mudge, A Farmer, T Paape, K Steel, GD Weiblen, A Bharti, P Zhou, GD May, ND Young, P Tiffin, Systematic Biology, 62(3): 424-438, DOI:10.1093/sysbio/syt009 (2013)
- “Selection, Genome Wide Fitness Effects, and Evolutionary Rates in the Model Legume Medicago truncatula,” T Paape, T Bataillon, P Zhou, T Kono, R Briskine, ND Young, P Tiffin, Molecular Ecology, 22(13):3525-3538, DOI:10.1111/mec.12329 (2013)
- “Estimating Heritability With Whole-Genome Data,” J Stanton-Geddes, J Yoder, R Briskine, ND Young, P Tiffin, Methods in Ecology and Evolution, DOI:10.1111/2041-210X.12129 (2013)
- “Candidate Genes and Genetic Architecture of Symbiotic and Agronomic Traits Revealed by Whole-Genome, Sequence-Based Association Genetics in Medicago truncatula,” J Stanton-Geddes, T Paape, B Epstein, R Briskine, J Yoder, J Mudge, AK Bharti, AD Farmer, P Zhou, R Denny, GD May, S Erlandson, M Sugawara, MJ Sadowsky, ND Young, P Tiffin, PLoS ONE, 8(5): e65688, doi:10.1371/journal.pone.0065688 (2013)
- “Whole-Genome Nucleotide Diversity, Recombination, and Linkage-Disequilibrium in the Model Legume Medicago truncatula,” A Branca, T Paape, P Zhou, R Briskine, AD Farmer, J Mudge, AK Bharti, JE Woodward, GD May, L Gentzbittel, C Ben, R Denny, MJ Sadowsky, J Ronfort, T Bataillon, ND Young, P Tiffin, Proceedings of the National Academy of Sciences of the USA, 108: E864-870, DOI:10.1073/pnas.1104032108 (2011)
- “The Medicago Genome Provides Insight Into The Evolution of Rhizobial Symbioses,” ND Young, F Debellé, G Oldroyd, R Geurts, SB Cannon, MK Udvardi, VA Benedito, KFX Mayer, J Gouzy, H Schoof, et al., Nature, 480: 520-524, DOI:10.1038/nature10625 (2011)
Note: Authors in bold are MSI Principal Investigators.
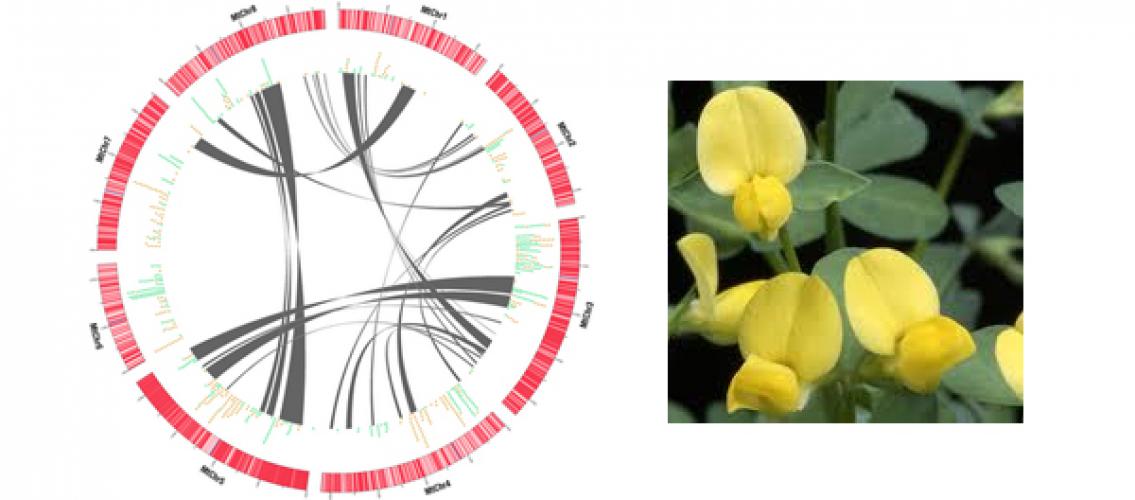
Image Description: Left: A standard circular plot of Medicago truncatula’s eight chromosomes. Lines are drawn between regions of the genome that have evidence of ancient duplication events, which are plentiful in this genome. On the outer ring, dots represent individual members of two large gene families known to have association with plant-microbe interactions. Right: Medicago truncatula (barrel medic).
Posted on November 27, 2013.