Above: Random events may tip the fate of living organisms. Master equations can be used to understand these random events.
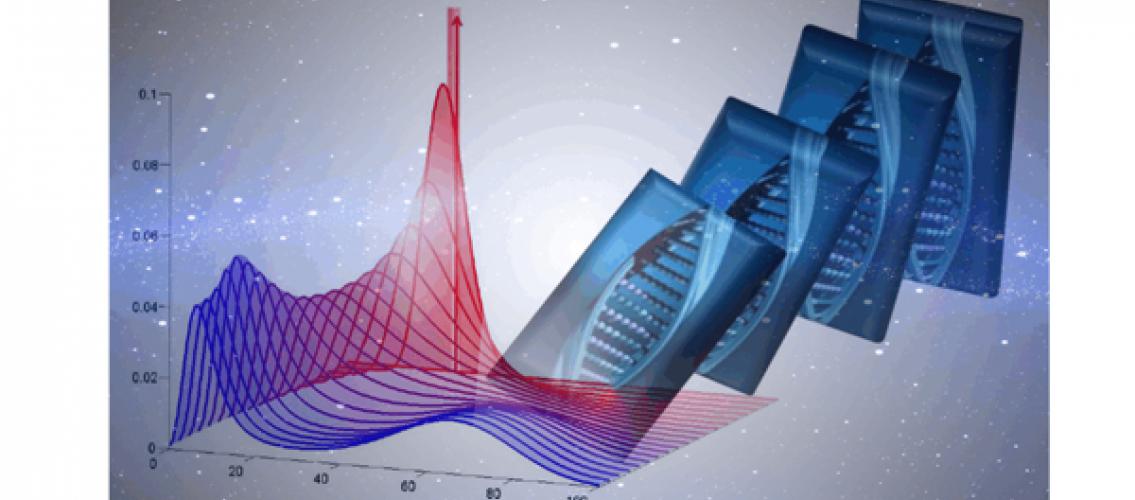
Biology is full of randomness – as random molecular events take place in organisms, populations evolve. Master probability equations can provide a complete model of this probabilistic behavior in biomolecular networks. These equations purport to govern all possible outcomes, hence the title “master.” Although these equations have enormous potential in biological research, their complexity has meant that solutions had only been found for the simplest molecular interaction networks.
MSI Principal Investigator and Fellow Yiannis Kaznessis and his PhD student Patrick Smadbeck recently published a paper that describes a numerical closure scheme for the master probability equation that governs random molecular events in chemical or biochemical reactions. The solution involves ordinary differential equations that describe the time evolution of probability distribution moments. The solution by Professor Kaznessis and Mr. Smadbeck will allow researchers to mathematically conceptualize a wide range of experimental observations. Their paper was published in the Proceedings of the National Academy of Sciences of the USA (“A Closure Scheme for Chemical Master Equations,” P Smadbeck, Y Kaznessis, PNAS, 110(35):14261, DOI: 10.1073/pnas.1306481110 (2013)).
Professor Kaznessis and his research group use MSI resources in their work to create new technologies that will be able to fight antibiotic-resistant bacteria. Microorganisms called enterococci have evolved over the years to resist almost all antibiotics, with the result that patients can develop serious infections that medical practitioners are unable to treat. The Kaznessis group uses molecular simulations to study biological interactions and functions.
Posted on January 8, 2014