Streptococcus suis is a global economic and welfare concern in the swine industry; pathogenic strains of S. suis are associated with central nervous system and systemic disease. Being able to differentiate pathogenic from commensal strains of S. suis easily and cost-effectively would be of great benefit to the swine production industry.
April Estrada, who was a 2020 UMII MnDRIVE PhD Graduate Assistant, worked on a project called “Genomic approaches to understand Streptococcus suis pathogenesis in the United States,” that used whole genome sequencing, statistical analyses, and relevant epidemiological data to determine virulence-associated genes associated with S. suis pathogenic strains in North America. She and her colleagues were able to complete two research goals with the UMII Graduate Assistantship:
- Identified two S. suis virulence-associated genes, ofs and srtF, that better identify pathogenic strains compared to the classical virulence-associated genes epf, mrp, sly of S. suis.
- Identified three novel candidate virulence-associated genes of S. suis by comparative pan-genomic analysis of 208 porcine S. suis isolates from North America.
This research contributes to the improvement of control and prevention strategies against S. suis.
Ms. Estrada is a member of the research group of MSI PI Connie Gebhart (professor, Veterinary and Biomedical Sciences) and used MSI resources for this project. The UMII MnDRIVE Graduate Assistantship program supports UMN PhD candidates pursuing research at the intersection of informatics and any of the five MnDRIVE areas:
- Robotics, Sensors and Advanced Manufacturing
- Global Food Ventures
- Advancing Industry, Conserving Our Environment
- Discoveries and Treatments for Brain Conditions
- Cancer Clinical Trials
This project is part of the Global Food Ventures MnDRIVE area. Publications resulting from this work include:
- Estrada A.A., M. Gottschalk, A. Rendahl, S. Rossow, L. Marshall-Lund, D.G. Marthaler, C.J. Gebhart. Proposed virulence-associated genes of Streptococcus suis isolates from the United States serve as predictors of pathogenicity. Porcine Health Management 7(1): 22 (2021). doi: 10.1186/s40813-021-00201-6.
- Estrada A.A., M. Gottschalk, C. Gebhart, D.G. Marthaler. Comparative analysis of Streptococcus suis genomes identifies novel candidate virulence-associated genes in North American isolates. Veterinary Research 53:23 (2022). doi: 10.1186/s13567-022-01039-8.
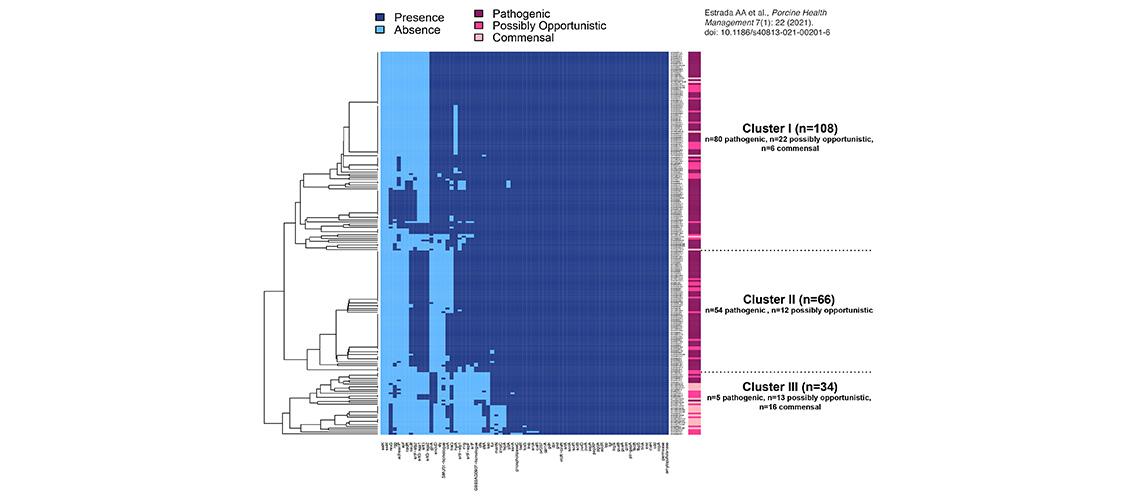
Image description: Virulence-associated gene (VAG) profiling of 208 S. suis isolates. Heatmap illustrating the presence and absence of 71 previously published VAGs in 208 isolates. Isolates are annotated (right) by pathotype (pathogenic, possibly opportunistic, commensal). Clustering of the 208 S. suis isolates by VAGs illustrated three clusters (Clusters I-III), two of which (Clusters II and III) suggest associations between VAGs and pathotype. Thirty-two (45%) VAGs were present in all genomes regardless of pathotype and were clearly not indicators of the pathogenic pathotype. Image and description modified from Estrada AA et al., Porcine Health Management 7(1): 22 (2021). doi: 10.1186/s40813-021-00201-6.
posted on November 8, 2022