New long-read sequencing technologies and bioinformatics pipelines have been developed that provide scientists with tools to more efficiently map and analyze genomes. Nanopore adaptive sampling (NAS), based on platforms pioneered by Oxford Nanopore Technologies, Inc., is being used by MSI PIs Peter Larsen (assistant professor, Veterinary and Biomedical Sciences) and Christopher Faulk (associate professor, Animal Science) to sequence the genomes of zoonotic pathogens and their hosts.
Professors Larsen and Faulk demonstrated the efficacy of using the NAS method in a project that selectively sequenced the DNA of bacterial pathogens in black-legged ticks (Ixodes scapularis) in the wild. Pathogens were able to be identified quickly, with little preparation, and in field conditions; since tick-borne pathogens are a serious health concern, these are important benefits. The research was published in July 2023: E.J. Kipp, L.L. Lindsey, B. Khoo, C. Faulk, J.D. Oliver, P.A. Larsen. Metagenomic surveillance for bacterial tick-borne pathogens using nanopore adaptive sampling. Scientific Reports 13: 10991 (2023). doi: 10.1038/s41598-023-37134-9.
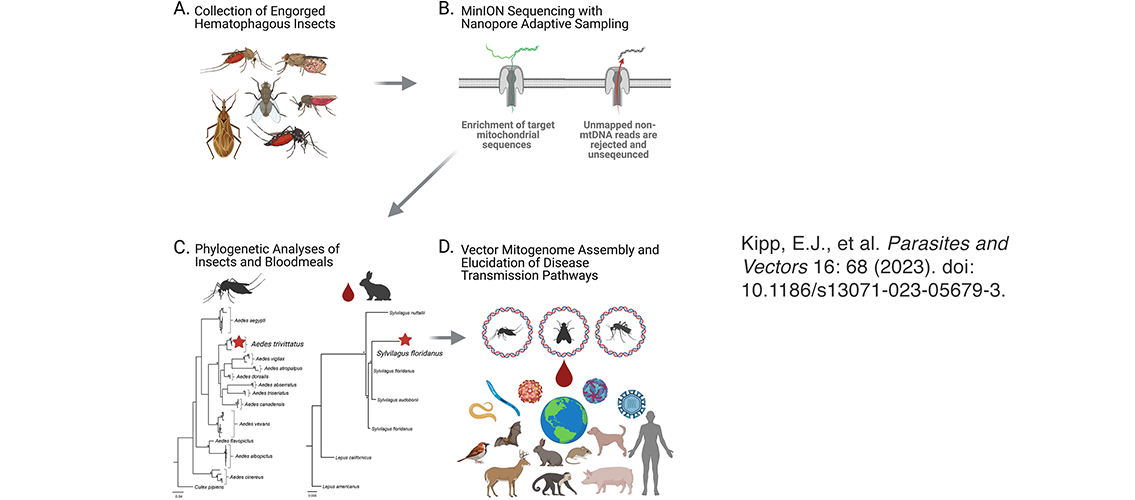
In another project, Professors Larsen and Faulk and their colleagues used NAS to sequence genomic DNA from four species of field-collected, blood-engorged mosquitoes (Aedes trivittatus, Aedes vexans, Culex restuans, and Culex territans) and one deer fly (Chrysops niger). They assembled and annotated complete mitochondrial genomes (mitogenomes) for these insects; compared to the data they generated with a control experiment that did not use NAS, the NAS experiment gave a substantially higher proportion of reference-mapped mtDNA reads. This streamlined the process of mitogenome assembly and annotation. The research was published in February 2023: E.J. Kipp, L.L. Lindsey, M.S. Milstein, C.M. Blanco, J.P. Baker, C. Faulk, J.D. Oliver, P.A. Larsen. Nanopore adaptive sampling for targeted mitochondrial genome sequencing and bloodmeal identification in hematophagous insects. Parasites and Vectors 16: 68 (2023). doi: 10.1186/s13071-023-05679-3.
These papers are part of the dissertation research of Evan Kipp (first author on both papers), who is a PhD candidate in the Larsen lab.
The researchers have also used NAS to sequence the genome of the endangered golden lion tamarin (Leontopithecus rosalia), a small primate found in South America. The research was published in the journal BMC Genomics in 2021: N. Wanner, P.A. Larsen, A. McLain, C. Faulk. The mitochondrial genome and epigenome of the golden lion tamarin from fecal DNA using nanopore adaptive sequencing. BMC Genomics 22: 726 (2021). doi: 10.1186/s12864-021-08046-7.
The Larsen lab uses high-throughput second and third-generation sequencing technologies available at MSI to investigate epigenomic vulnerabilities contributing to the origin of sporadic disease and emerging zoonoses at the wildlife-livestock interface. The Faulk lab investigates functional genomics with nanopore sequencing data and comparisons of RNA-seq data, bisulfite sequencing, and reduced representation bisulfite sequencing; they use Galaxy and other resources provided by MSI.
Image description: Graphical abstract of NAS analysis of mosquitoes and flies.
posted on September 13, 2023