Macrophages are important immune cells that detect and destroy invading cells. The macrophages that reside in the mammary gland are also important for mammary gland development and tissue homeostasis. MSI RIS staff are working with researchers in the Department of Laboratory Medicine and Pathology (LM&P) to study these cells.
A recent paper used transcriptional profiling and single-cell RNA sequencing approaches to identify a distinct macrophage subpopulation residing in mouse mammary glands. This macrophage subpopulation was distinct from other macrophages because of Lyve-1 expression. Lyve-1 is a receptor for the extracellular matrix component hyaluronan. This finding indicates that these macrophages can influence the extracellular environment within the mammary gland tissue, which includes not only cells but also but also a complex and rich mixture of extracellular proteins, sugars, and minerals. Both the cell and extracellular components in the tissue are impacted by tissue-resident macrophages. Further understanding of macrophage function in the tissue will help researchers understand how macrophages might contribute to tissue-specific disease such as breast cancer.
Co-authors on this paper include Professor Kaylee Schwertfeger and Associate Professor Andrew Nelson, both MSI PIs in the LM&P department, and RIS analyst Dr. Rebecca LaRue. Dr. LaRue analyzed the single-cell RNA sequencing that included differential gene expression analysis and pathway analysis. The paper can be found on the journal website: Y Wang, TS Chaffee, RS LaRue, DN Huggins, PM Witschen, AM Ibrahim, AC Nelson, HL Machado, KL Schwertfeger. 2020. Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. Elife. Jun 1;9:e57438. doi: 10.7554/eLife.57438.
Another paper co-authored by Professors Schwertfeger and Nelson was featured on the MSI website in August 2020: Improving Treatments for Breast Cancer.
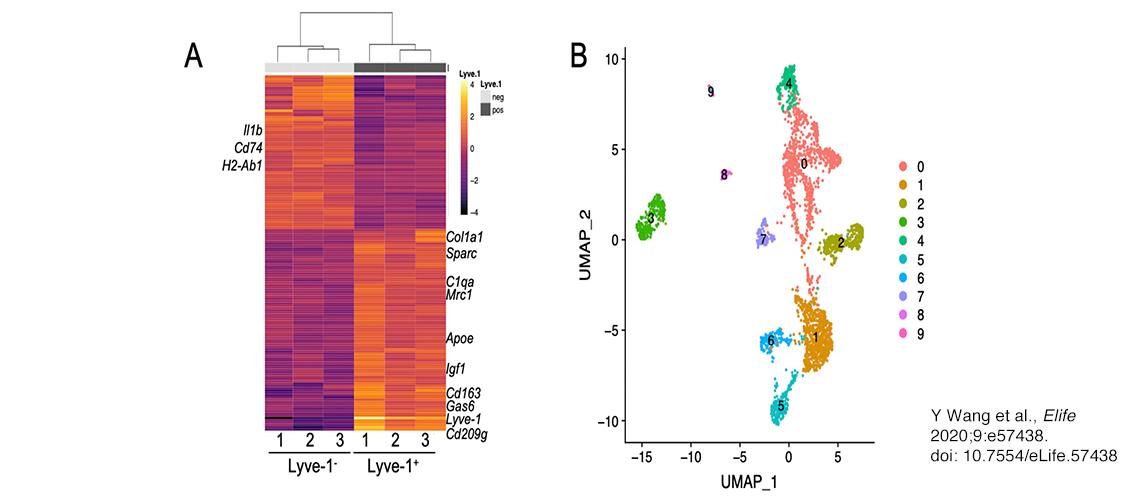
Image description: Identification of a distinct Lyve-1+ macrophage subpopulation by transcriptional profiling. (A) Heat map of RNA-seq analysis of CD45+CD11b+F4/80+Lyve-1– and CD45+CD11b+F4/80+Lyve-1+ cells isolated from 6-week-old (lane 1) and 10-week-old (lanes 2–3) mice. Genes shown have an adjusted p-value <0.01 and a fold-change of >1 and <10 (abs values). For each lane, n = 4 pooled mice. (B) UMAP (Uniform Manifold Approximation and Projection) of scRNA-seq analysis of CD45+ cells isolated from 10-week-old mice as generated by Seurat software. Image and description from Y Wang et al., Elife 2020;9:e57438. doi: 10.7554/eLife.57438.
posted on August 17, 2021