Invasive plant species are a threat to native species in many habitats. Researchers are developing computational models that will allow them to understand the conditions that allow the spread of non-native plants and how to predict it. MSI users in the College of Biological Sciences have developed and evaluated several species distribution models (SDMs) based on known facts about the spread of the invasive species Palmer amaranth (Amaranthus palmeri). This plant has spread from its native habitat in the desert of the US southwest and Mexican northwest into much of the US. The study authors are MSI PIs David Moeller (associate professor, Plant and Microbial Biology) and Peter Tiffin (professor and head, Plant and Microbial Biology), Dr. Ryan D. Briscoe Runquist (post-doc, Moeller lab and lead author of the study), and Thomas Lake (PhD student, Moeller lab).
The study, published in February 2019 in the journal Scientific Reports, used detailed data about the spread of A. palmeri during 1970 through 2017 to test the ability of SDMs to accurately predict how it might continue to spread. The researchers were able to make several interesting conclusions about these models. For example, models based on more recent data performed better at predicting how the plant would spread into new areas, and the researchers found that models that used only conditions in newly invaded areas better predicted spread than those that also included conditions in the plant’s native range. The study can be read on the Scientific Reports website: Ryan Briscoe Runquist, Thomas Lake, Peter L. Tiffin, David Moeller. 2019. Species distribution models throughout the invasion history of Palmer amaranth predict regions at risk of future invasion and reveal challenges with modeling rapidly shifting geographic ranges. Scientific Reports 9(1). doi: 10.1038/s41598-018-38054-9.
Following on the success of the models of Palmer amaranth spread, the researchers are expanding their research to create models for other invasive species. They are currently working on a paper that will describe predictive model maps of eight additional invasive species. The group also has a project to develop models that use machine-learning and deep-learning methods, drawing on the computational power available with high-performance computers. This project will use high-resolution satellite images to train deep-learning algorithms to identify the invasive species leafy spurge and common tansy in the images. These algorithms will then be able to identify new populations and quantify population sizes and population growth rates of these species.
Professor Moeller’s research group uses MSI for studies of the ecological and genetic mechanisms of speciation as well as the ecology and evolution of plant reproductive systems, including plant-pollinator interactions and plant mating systems. Professor Tiffin’s group uses MSI for genomics studies of bacteria that form symbiotic relationships with various legumes and the plant pathogen Aphanomyces. They are also working on RNA-seq analyses of several plant species.
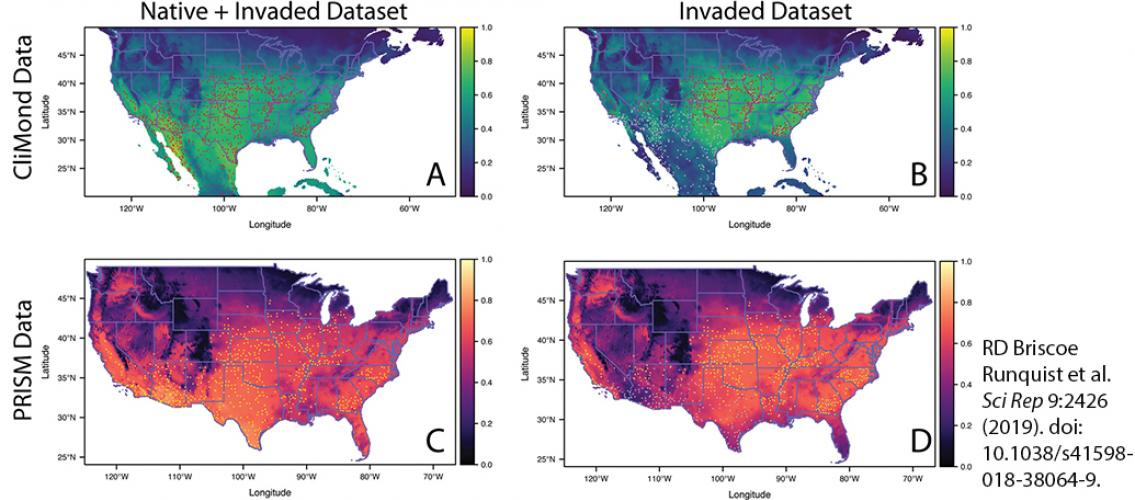
Image description: SDM model projections of Amaranthus palmeri generated using variables from the climate datasets CliMond and PRISM for the native + invaded dataset and invasive-only dataset. All projections are means of 25 model runs. Panels A and B were generated using CliMond environmental variables (blue/green color palette) and panels C and D were generated using PRISM variables (pink/purple color palette). Panels A and C used the native + invaded dataset for model building and panels B and D used the invasive only dataset. In panels A and B the occurrences included in the model are shown in red and in panels C and D the occurrences included in the model are shown in yellow. All other occurrences are in gray. The complementary log-log predicted probability for each raster cell is indicated by the color corresponding the bars to the right of the panels. Image and description adapted from R.D. Briscoe Runquist et al. Scientific Reports 9:2426 (2019). doi: 10.1038/s41598-018-38054-9.
posted on July 31, 2019