Application of Informatics to Transcription of Ancient Papyri
While computers can do many things, there are still a few areas in which humans excel such as the discriminatory power of the eye and the natural human ability to quickly classify objects. The visual ability of recognizing patterns is at the core of the Zooniverse (https://www.zooniverse.org/) citizen science project that Professor Lucy Fortson (School of Physics and Astronomy, College of Science and Engineering) has been involved with. It started with Galaxy Zoo in 2007 by simply asking the general public to help classify about a million scientific images of galaxies and since has grown to over 25 projects enlisting the help of the public to identify whales, lions, and even planets outside our solar system.
As part of an interdisciplinary team, MSI staff have been working with Professor Fortson and her Humanities colleagues at the University of Minnesota and Oxford University (UK) to help transcribe a collection of ancient papyri. The papyri are part of the Oxyrhynchus collection maintained by Oxford University and composed of over 500,000 fragments dating from the period 150 BCE to 650 CE and excavated from the ancient trash heap of the Egyptian town called Oxyrhynchus. Contributors to the Ancient Lives (http://www.ancientlives.org/) citizen science project, members of the general public, are asked to help transcribe the contents of these individual papyri. No ancient or foreign language skills are required as the project relies solely on visual pattern recognition. Volunteers are simply asked to match characters on the papyrus to corresponding characters on an electronic virtual keyboard by first clicking the letter on the papyrus image and then clicking the corresponding Greek letter. As every single papyrus will be transcribed by many different users, a consensus will emerge from the many transcriptions. As of November 2014, nearly ten million marks have been made on over 150,000 fragments by about a million volunteers worldwide.
This wealth of clicks needs to be turned into a data product useful to the Humanities researchers through the development of a data processing pipeline. This is where Professor Fortson’s background in astrophysics and the MSI team come in. While there are many steps in the pipeline, one of the most critical is the consensus algorithm. Applying kernel density estimation (KDE) methods to the volunteers’ contributed transcription data, MSI staff developed a workflow that converts clicks into computationally deducted consensus sequences, or text strings, and thus quickly enabled the transformation of physical documents into computationally searchable data.
To enable the organization of these data sets, MSI has also developed an editorial web tool (http://papyrus.msi.umn.edu/) to support the curation and metadata annotation efforts of these data sets by scholars of ancient texts. In a final step MSI staff and collaborators at Middle Tennessee State University are applying bioinformatics tools to identify words or text strings and similarities between papyri (e.g. copies of known texts).
With a 2013 award from the National Endowment for the Humanities, the team, now led by Drs. Philip Sellew and Nita Krevans, University of Minnesota professors in Classical and Near Eastern Studies, is applying a similar strategy to the transcription of ancient Coptic papyri.
A new initiative, Zooniverse@UMN, has recently been funded* to support University of Minnesota-affiliated projects. This effort is currently soliciting proposals for text-based projects that would benefit from hundreds of thousands of online volunteers transcribing or metadata tagging a digitally imaged collection. Researchers can download the Request for Proposals on the UM Zoomanities webpage. The proposal due date window is November 24 - December 15, 2014.
Publications by these researchers include:
- Williams, A.C., Wallin, J.F., Yu. H, Carroll, H.D., Lamblin., A-F., Fortson, L., Obbink, D., Lintott, C.J. & Brusuelas, J.H. (2014). A Computational Pipeline For Crowdsourced Transcriptions of Ancient Greek Papyrus Fragments. (To Appear In) Proceedings of the 2nd Workshop on Big Humanities Data.
- Williams, A.C., Carroll, H.D., Wallin, J.F., Brusuelas, J., Fortson, L., Lamblin., A-F., & Yu, H. (2014). Identification of Ancient Greek Papyrus Fragments Using Genetic Sequence Alignment Algorithms. (To Appear In) Proceedings of the 1st Workshop on Digital Humanities and e-Science.
*Funding for Zooniverse@UMN is provided by the Office of the Vice President for Research, the University Libraries, the Colleges of Biological Sciences, Liberal Arts, and Science and Engineering, and the University of Minnesota Informatics Institute.
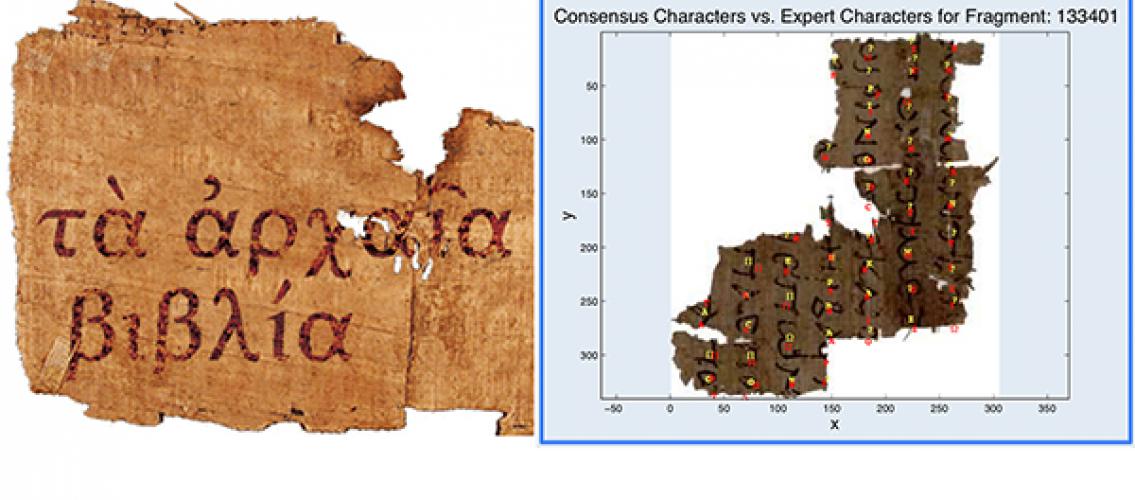
Figure descriptions: Left: a fragment from the Oxyrhynchus papyri. Right: an example of a transcribed fragment plotted on the image of the original fragment. Yellow characters are the consensus characters for the volunteers who transcribed the fragment, while the red characters are the transcription of a Greek expert. The expert characters have been shifted down a bit to provide better readability. All users’ transcriptions for the fragment are also kept in a text file for Greek scholars to review.
posted on November 26, 2014