Fusion genes are genes made by joining parts of two different genes. They are implicated in the development of some kinds of cancer because they can produce more abnormal proteins than normal genes. The precursors to these proteins are fusion transcripts, a type of RNA. Certain fusion genes and their associated proteins are connected with specific types of cancer.
MSI PIs David Largaespada (professor, Pediatrics; Genetics, Cell Biology, and Development; Masonic Cancer Center; Center for Genome Engineering) and Branden Moriarity (assistant professor, Pediatrics; Masonic Cancer Center; Center for Genome Engineering) and colleagues recently authored a paper describing a new pipeline for finding fusion transcripts. They developed a pipeline that combines existing software with a custom-developed program and used it to evaluate RNA-sequence data from osteosarcomas in humans and dogs. The pipeline includes tools available in MSI’s Galaxy instance.
The new pipeline allowed the researchers to identify neoantigens that are associated with fusions. With this information, it may be possible to develop treatments that are targeted to the specific tumor. The researchers also discovered more similarities between canine and human forms of osteosarcoma than were previously known. This is helpful because dogs develop osteosarcoma more often than humans do, and similarities between the forms of cancer may allow drug developers to test vaccines on dogs prior to human trials. The paper was published in the journal Scientific Reports in early 2019: SK Rathe, FE Popescu, JE Johnson, AL Watson, TA Marko, BS Moriarity, JR Ohlfest, DA Largaespada. 2019. Identification of Candidate Neoantigens Produced by Fusion Transcripts in Human Osteosarcomas. Scientific Reports 9:358. DOI: 10.1038/s41598-018-36840-z.
Professor Largaespada’s group uses a transposon system called Sleeping Beauty for insertional mutagenesis for cancer gene discovery and uses next-generation sequencing tools available through MSI. Dr. Sue Rathe and Dr. Flavia Popescu are MSI users and post-docs in the Largaespada group. Professor Moriarity’s research group uses MSI resources for studies of the genetics of pediatric cancer in order to develop new treatments. JJ Johnson is a Senior Software Developer in MSI’s Application Development Solutions group. Dr. John Ohlfest, who was also an MSI PI, was instrumental in planning the initial mouse experiments and establishing the long-term goals for this project. He passed away in 2013.
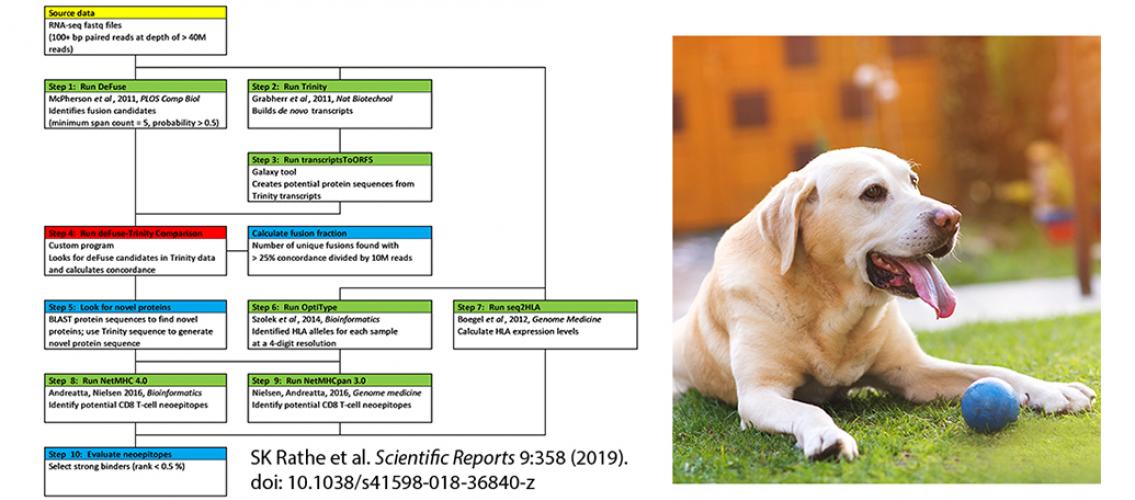
Image description: Left: Diagram of the methodology used to identify potential neoantigens in OS. Starting data highlighted in yellow, pre-existing programs highlighted in green, custom programs highlighted in red, and manual processes highlighted in blue. Image and description from SK Rathe et al. Scientific Reports 9:358 (2019). doi: 10.1038/s41598-018-36840-z Right: Osteosarcoma is more common in dogs than in humans, and there are similarities between canine and human types of the disease.
posted on May 20, 2019