Wheat leaf rust is caused by a fungus (Puccinia triticina) that attacks wheat and other cereal crops. The fungus is found in wheat populations worldwide and is a leading cause of yield loss. Agricultural researchers are studying the genetics of P. triticina with the goal of developing wheat varieties that are resistant to it.
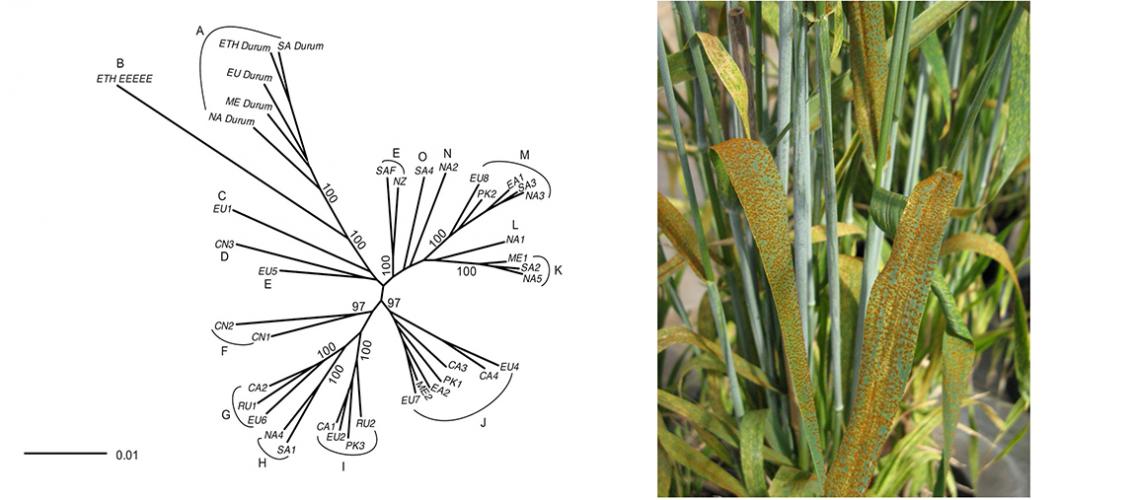
MSI PI James Kolmer (USDA - ARS Cereal Disease Lab; adjunct professor, Plant Pathology) is leading a research project that involves analyzing worldwide populations of the wheat leaf rust fungus Puccinia triticina for genetic variation. In a paper published in late 2019 in the journal Heredity, Professor Kolmer and colleagues, including Dr. Adam Herman from MSI’s Research Informatics Solutions group, performed sequence-by-genotyping analyses in order to determine the genetic relatedness of different populations of P. triticina. The analysis included 558 isolates of the fungus from wheat-producing regions in North America, South America, Europe, the Middle East, Ethiopia, Russia, Pakistan, Central Asia, China, New Zealand, and South Africa. The results showed genetic similarities between varieties from neighboring areas, plus indications of migration between regions in varieties that were physically more distant but genetically similar. The paper can be found on the journal website: JA Kolmer, A Herman, ME Ordoñez, S German, A Morgounov Z Pretorius, B Visser, Y Anikster, M Acevedo. 2019. Endemic and panglobal genetic groups, and divergence of host-associated forms in worldwide collections of the wheat leaf rust fungus Puccinia triticina as determined by genotyping by sequencing. Heredity 124:397–409. doi: 10.1038/s41437-019-0288-x.
Related research by the Kolmer group was featured on the MSI website in June 2016: Identifying Rust Resistance Genes in Wheat.
Image description: Left: Neighbor joining plots of 556 Puccinia triticina isolates in 40 MLG genotype groups in 11 regional populations. NA North America; SA South America; EA East Africa; ME Middle East; NZ New Zealand; SAF South Africa; EU Europe; PK Pakistan; CN China; CA Central Asia; RU Russia. Plot of Prevosti’s genetic distance of isolates in MLG groups based on 6745 SNP markers. Indicated clusters (A-O) of isolate groups have support values over 95% based on 1000 bootstrap samples. Image and description, JA Kolmer, et al. Heredity 124:397–409 (2019). doi: 10.1038/s41437-019-0288-x. Right: Wheat flag leaf covered by rust infections. Photo from James Kolmer.
Posted on November 17, 2020