When patients suffer renal failure, a kidney transplant is the most cost-effective treatment. Unfortunately, in spite of modern anti-rejection drugs, the patients sometimes suffer either acute rejection (AR), which can happen up to a year or so after the transplant, or chronic allograft dysfunction (CGD), in which the transplanted kidney gradually loses function over a longer time.
Several MSI Principal Investigators (PIs) from the School of Public Health, College of Pharmacy, and Medical School and members of their research groups have recently published findings where they used RNA-seq analysis to identify changes in gene expression after transplants. The researchers hypothesize that gene expression changes after immunosuppressive drugs are started and continues to do so following the transplant. This analysis is a first step towards the eventual goal, which is to be able to find the optimal genetic signature for immune-system suppression. This will allow doctors to create individualized immunosuppressive treatments that will successfully avoid AR and CGD.
The paper was published last year in the journal PLoS One: C. Dorr, B. Wu, W. Guan, A. Muthusamy, K. Sanghavi, D.P. Schladt, J.S. Maltzman, S.E. Scherer, M.J. Brott, A.J. Matas, P.A. Jacobson, W.S. Oetting, and A. Israni. 2015. Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PloS One 10(5) (MAY 06), 10.1371/journal.pone.0125045. The MSI PIs among the co-authors are:
- Assistant Professor Weihua Guan, Biostatistics
- Associate Professor Ajay Israni, Medicine, Adjunct Epidemiology and Community Health
- Professor Pamala Jacobson, Experimental and Clinical Pharmacology
- Professor William Oetting, Experimental and Clinical Pharmacology
- Associate Professor Baolin Wu, Biostatistics
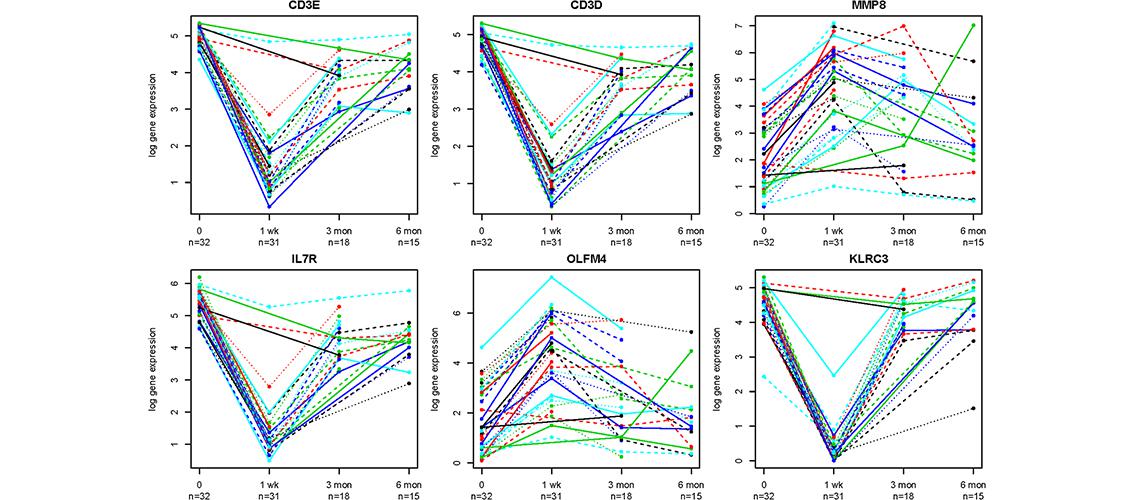
Image description: Expression of representative genes over time in kidney transplant recipients. Representative time series of fold expression changes relative to baseline (time 0) of some top genes with higher and lower level compared to baseline. Each line on each graph represents the expression of the particular gene in a separate kidney allograft recipient. Note that all patients do not have data all the time points. CD3E = CD3 Epsilon TCR complex; CD3D = CD3 Delta TCR complex; MMP8 = Matrix Metallopeptidase 8; IL7R = Interleukin 7 Receptor; OLFM4 = Olfactomedin 4; KLRC3 = Killer Cell Lectin-like Receptor subfamily C, member 3. Image and description: C Dorr et al. PloS One 10(5), 10.1371/journal.pone.0125045 (2015).
posted on February 17, 2016