Fowlpox is a viral disease of chickens and turkeys that occurs worldwide. Fowlpox viruses contain an enzyme called a resolvase, which allows the virus to cleave a DNA strand in particular 3D structures. The fowlpox virus resolvase is able to do this at any part of its genome, rather than only at the site of a specific DNA sequence.
In a recent paper, MSI PI Hideki Aihara (associate professor, Biochemistry, Molecular Biology, and Biophysics) and colleagues revealed the crystal structure of fowlpox virus resolvase. This research helps explain how the fowlpox virus is able to cleave DNA in structure-specific, but sequence non-specific fashions. The paper can be read on the website of the journal Scientific Reports: N Li, K Shi, R Rao, S Banerjee, H Aikara. 2020. Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase. Scientific Reports 10(1). doi: 10.1038/s41598-019-56825-w.
Professor Aihara uses MSI resources to analyze X-ray crystallographic images to determine the 3D structures of protein-DNA complexes as part of studies into DNA recombination, repair, and mutation processes relevant to human health. Co-author Ke Shi uses MSI resources as part of the Aihara research group. They recently participated in a project to describe SARS-CoV-2, the coronavirus that causes COVID-19.
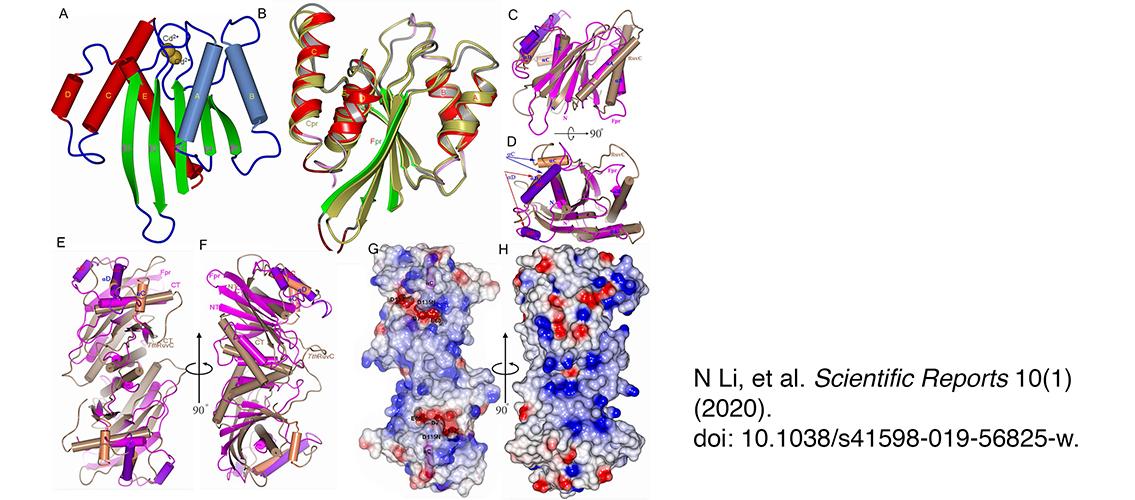
Image description: Overall structure of the fowl poxvirus resolvase (Fpr). (A) Structure of Fpr with the secondary structure elements labeled. (B) Superposition of Fpr and Cpr monomers based on the central five β-strands. (C) Superposition of Fpr and RuvC from T. thermophiles (TthRuvC) monomers, highlighting that two proteins have different αC and αD helices (colored differently from the rest of the monomers and indicated by arrows). αB and the preceding loop in TthRuvC are longer than the counterparts in Fpr, while αC in Fpr superposes onto αD of TthRuvC but is much longer. (D) 90° rotation of C around a horizontal axis. (E,F) Superposition of Fpr and RuvC dimers showing the different shape of Fpr and TthRuvC, F is 90° rotation of E around a vertical axis. (F) Electrostatic surface representation of the Fpr dimer. The negative surface (in red) formed by the active site residues can be seen at the bottom of the DNA binding grooves. The αC helices are shown in magenta. (H) 90° rotation of G around a vertical axis, showing strong positive potentials (blue) on the side of active site of the Fpr dimer. The orientation of G and H are the same as E and F respectively. Image and description, N Li, Scientific Reports 10(1) (2020), doi: 10.1038/s41598-019-56825-w.
posted on June 22, 2020