Cells move, or migrate, across body tissues such as bone, brain, muscle, and fat. This migration is how wounds heal, for example. Unfortunately, it is also the mechanism by which cancer spreads. Different types of tissues have varying degrees of stiffness, and each kind of cell has a preferred stiffness for migration. The scientists who study cell migration refer to the cell’s “engine,” the mechanism that generates force in the cell, and the “clutch,” which is made of integrins (a kind of protein) that transfers the force from the cell to the surrounding environment. A fast-spinning engine needs a stiffer surface on which to move.
MSI Principal Investigator David Odde (professor, Biomedical Engineering) and his colleagues have developed a mathematical model that simulates how cells move through tissues of various stiffnesses. The model, which has been run on MSI’s supercomputers, is the subject of a recent paper in the journal Nature Communications. By using the model, the researchers are able to determine the optimal stiffness of surrounding tissues for cell migration. They can then stop migration by targeting the cell with drugs that change the speed of the engine-and-clutch system so that it can’t move well on the surrounding tissue.
The paper is available on the journal website (authors in bold are MSI users): BL Bangasser, GA Shamsan, CE Chan, KN Opoku, E Tuzel, BW Schlichtmann, JA Kasim, BJ Fuller, BR McCullough, SS Rosenfeld, and DJ Odde. 2017. Shifting the optimal stiffness for cell migration. Nature Communications 8:15313. DOI: 10.1038/ncomms15313.
Professor Odde is a principal investigator in the University’s Physical Sciences Oncology Center, which was funded last year with a $8.2 million grant from the National Cancer Institute. The purpose of the Center is to use physical sciences to help understand the behavior of cancer cells. An article about the grant appears on the University of Minnesota News page: U awarded multimillion-dollar grant for physics-based approach to cancer. The Center was also featured in the Spring 2017 issue of the U of M Foundation’s Legacy magazine: Predicting cancer's next move.
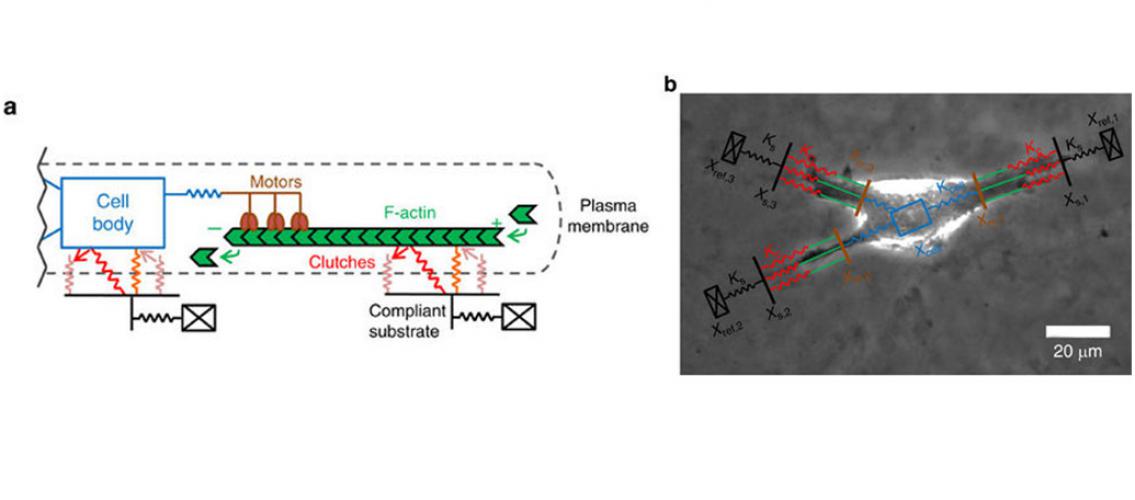
Image description: a) Schematic of a motor-clutch module attached to the central cell body. Additional modules may also extend from the cell body but are not shown here for simplicity. b) Representative schematics of the cell migration simulation overlaid on top of a phase-contrast image of U251 glioma cell. This image demonstrates how the simulator captures the three main protrusions of the cell. Image and description adapted from BL Bangasser et al. NComms 8:15313 DOI: 10.1038/ncomms15313.
posted on August 7, 2017