One of the most promising methods of treating cancer is called “tumor-homing” or “targeted drug” therapy. In this method, a cancer drug is linked to a kind of peptide called a “tumor-homing” peptide – the tumor-homing peptide recognizes a specific receptor that appears on tumors, thereby guiding the drug directly to the tumor. One such receptor that has shown promise for targeted drug therapy is called aminopeptidase N (APN).
MSI PI Fang Li, an associate professor in the Department of Pharmacology (Medical School), and his group recently published a paper in the Journal of Biological Chemistry in which they investigated APN-based targeted drug therapy. APN has two unusual functions: it is a receptor for tumor-homing peptides and also mediates cancer cell motility. Using crystallographic and biochemical methods, the authors studied both these functions and found that the same mechanism drives both of them. This research has implications for the development of cancer therapies that target APN.
The Li group uses MSI for studies of the structural and molecular basis of human diseases. The project described in this paper used computer resources available through MSI’s Basic Sciences Computing Laboratory. You can read the entire paper on the Journal of Biological Chemistry website (Chang Liu, Yang Yang, Lang Chen, Yi-Lun Lin, and Fang Li. A unified mechanism for aminopeptidase N-based tumor cell motility and tumor-homing therapy. The Journal of Biological Chemistry 289, (50) (DEC 12), 10.1074/jbc.M114.566802.
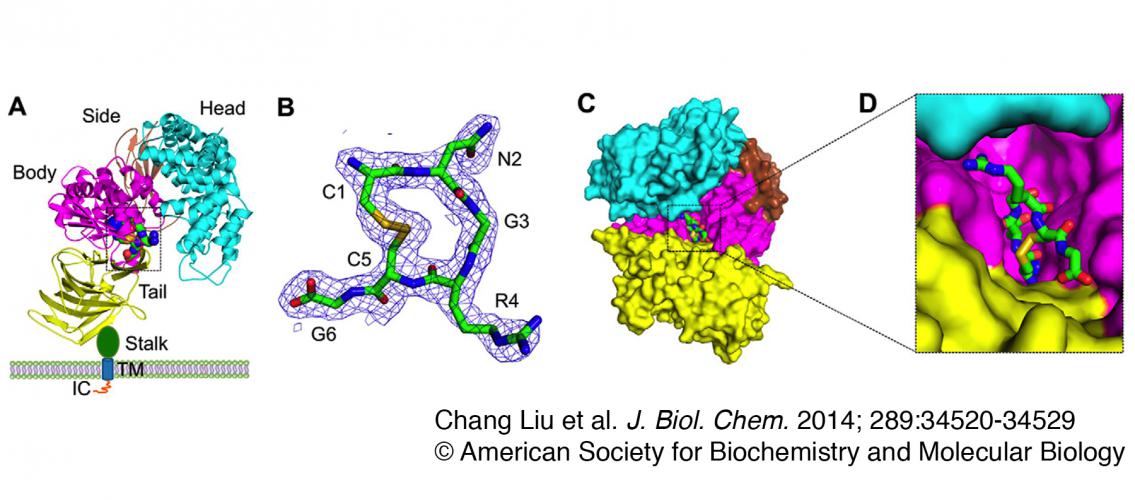
Image Description: Crystal structure of porcine APN in complex with a tumor-homing peptide CNGRCG A, overall structure of the pAPN-CNGRCG complex. pAPN contains an ectodomain, a stalk, a transmembrane anchor (TM), and an intracellular tail (IC). The ectodomain contains four domains: head (cyan), side (brown), body (magenta), and tail (yellow). CNGRCG is shown in green as balls and sticks. Zinc is shown as a blue ball. Only one monomer of the dimeric pAPN is shown. B, electron density map of CNGRCG. The electron density map corresponds to Fo − Fc omit map calculated in the absence of CNGRCG and contoured at 2.2 σ. C, another view of the pAPN-CNGRCG complex. The view of the complex is obtained by rotating the view in A first by 90° along a vertical axis and then by 45° along a horizontal axis, in such a way that the active site cavity of pAPN is facing the reader. D, enlarged view of CNGRCG in the active site of pAPN. Image and description from Chang Liu et al., J. Biol. Chem. 2014. 289:34520-34529. © 2014 American Society for Biochemistry and Molecular Biology.
posted on June 10, 2015