Breast cancer is a leading cause of death for women. Researchers know that genetics play an important part in one’s chance of getting the disease, but there is still much that they don’t know. Genome-wide association studies have identified a large number of genetic variants linked with the disease risk, but those still account for only about one-third of genetic contributions to breast cancer.
The research group of MSI PI Chad Myers (associate professor, Computer Science and Engineering; Masonic Cancer Center) is using MSI for several projects that use computational methods to study how combinations of genes result in breast cancer. The group hypothesizes that the interaction of genetic variants could play a role in cancer risk. Unfortunately, testing all the possible combinations would be extremely difficult, both statistically and computationally. To solve this problem, the Myers group developed a computational method called BridGE, which leverages the tendency of genetic interactions to cluster into coherent groups, thereby significantly reducing the number of interactions that must be tested.
BridGE was one of two grand prize winners in the “Up for a Challenge (U4C) Breast Cancer Challenge Award” held by the National Cancer Institute. The researchers published this work in the journal PLoS Genetics in September 2017. The paper appears on the PLoS Genetics website: Pathway-Based Discovery of Genetic Interactions in Breast Cancer.
The lead author of the paper, Wen Wang, used this research for a poster she presented at the 2017 MSI Research Exhibition. She was the Grand Prize winner in the Biological and Medical Sciences division in the Exhibition poster competition, receiving a travel award of $2,000. Dr. Wang was a PhD student when she worked on this project; she is now a post-doc in the Myers group.
Another MSI PI, Carol Lange (professor, Medicine; Masonic Cancer Center) collaborates with the Myers group and is a co-author on this study. Professor Lange uses MSI for research into the role of progesterone receptors in breast cancer.
Professor Myers and his research were recently featured on the College of Science and Engineering website: Chad Myers: Exploring genetic connections in breast cancer. Current research projects are also described on their lab’s website.
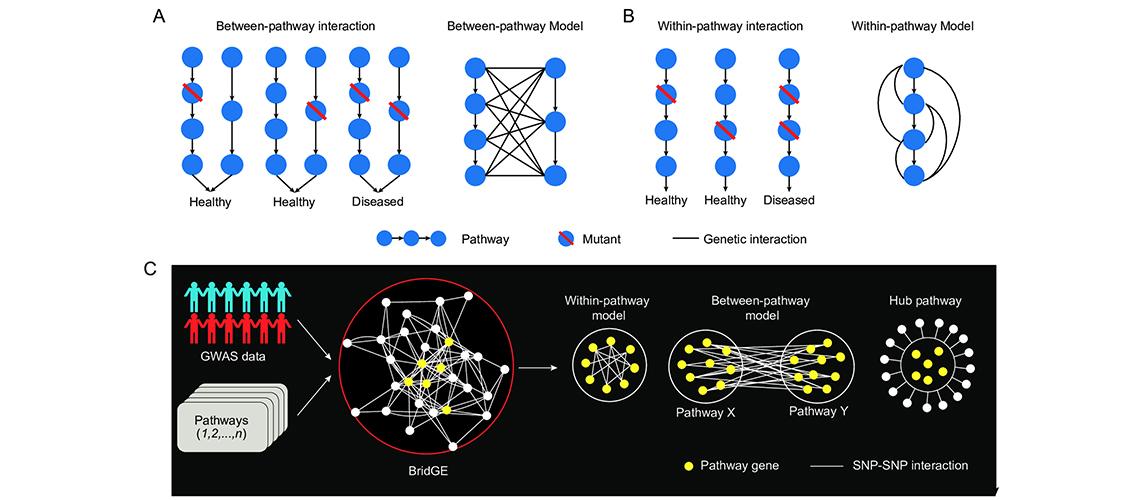
Image description: Pathway-level genetic interaction models. (A) Between-pathway interaction and between-pathway model. Two biological pathways share a common function necessary for maintaining a healthy state. Genetic variants in individual pathways do not result in a phenotype, but joint mutations in both pathways in the same individual results in disease. Between-pathway interactions clustering between two complementary pathways and appear are referred to as an instance of the between-pathway model (BPM). (B) Within-pathway interaction and within-pathway model. A single pathway supports a function for maintaining a healthy state. A single genetic variant does not result in a phenotype, but joint mutations in the same pathway results in the loss of function and a disease state. Within-pathway interactions clustered within the single pathway are called a within-pathway model (WPM). (C) Overview of the framework for discovering pathway-level genetic interactions from GWAS breast cancer data, leveraging the BridGE method. Image and description, Wang W, et al. PLoS Genet 13(9): e1006973 (2017).
posted on September 5, 2018