Research has shown that the human microbiome - the community of microorganisms that inhabit our bodies - may have health implications. It’s possible, for example, that a certain mixture of these microorganisms may cause certain diseases.
Researchers have recently begun to study the microbiome in depth. While much recent research has involved the bacteria that live in human bodies, only limited studies have dealt with the fungi that are also present. While there are fewer fungi present in the microbiome than there are bacteria, fungi also affect human health. For example, fungi cause some life-threatening diseases in infants, and other fungi may alleviate intestinal inflammation.
In a recent paper in PLoS One, MSI Principal Investigators Cheryl Gale (Associate Professor of Pediatrics and Genetics, Cell Biology, and Development), Dan Knights (Assistant Professor of Computer Science and Engineering), and Michael Sadowsky (Director, BioTechnology Institute; Professor of Soil, Water, and Climate) describe a new strategy to characterize intestinal fungal communities in infants. The authors used genomic techniques to study and characterize the fungal microbiomes of infants. They also obtained quantitative information about Candida, which is the dominant fungal genus in the human intestine. This work used software available through MSI as well as computational resources. The article can be read on the PLoS One website: Timothy Heisel, Heather Podgorski, Christopher M. Staley, Dan Knights, Michael J. Sadowsky, and Cheryl A. Gale. 2015. Complementary Amplicon-Based Genomic Approaches for the Study of Fungal Communities in Humans. PLoS One 10(2): e0116705. 10.1371/journal.pone.0116705.
Professors Gale, Knights, and Sadowsky use MSI resources to support their research into microbes and the microbiome. Professor Gale is using genomic approaches to establish how intestinal microbiome dynamics are associated with diseases in infants. Professor Knights is helping to create a bioinformatics pipeline for linking genetic variation with bacteria. Professor Sadowsky studies the metagenome of the Mississippi River, soil, and the human intestinal tract.
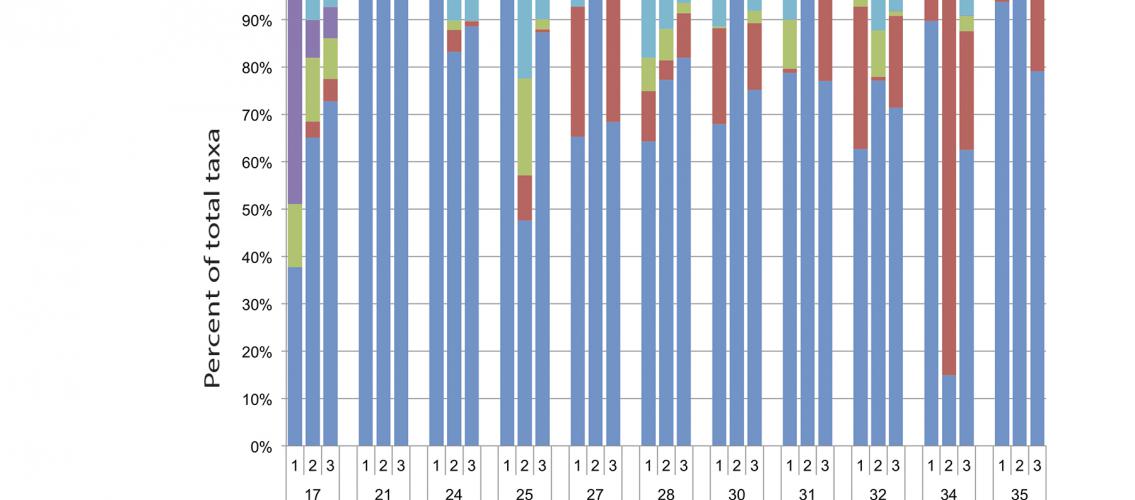
Image description: Relative Abundances of fungal taxa in infant fecal samples. Sample numbers (each from different infants) are noted on the lower line of the x-axis and the results of triplicate determinations are shown above the sample number for each. The most abundant taxa are indicated on the right-hand side of the graph, with low-abundance (present at <1.5% mean abundance across all samples) being grouped into the “Other” category. Sequencing reads are displayed as percentages of the total number of reads for each individual sequencing replicate. Image and description from Heisel, T, et al., 2015. PLoS One 10(2): e0116705. 10.1371/journal.pone.0116705.
posted on April 15, 2015