The microbiome of the human digestive tract has become an area of a great deal of exciting research and discovery. Scientists are finding out how the microorganisms living in our bodies affect our health, and several MSI PIs are involved in this research.
MSI Principal Investigators Timothy Starr, Dan Knights, and Ran Blekhman are co-authors on recently published research that describes the discovery of specific changes in the microbial community caused by colorectal cancer tumors. The researchers found increased levels of various microbes, especially including the genus Fusobacterium, which has been identified previously as being related to colorectal cancer, and the genus Providencia. The latter genus has not been identified before as having these pathogenic qualities. The study also showed that there is an increase in virulence-associated genes in the colorectal cancer microenvironment. The paper was published in the journal Genome Medicine in June (MB Burns, J Lynch, TK Starr, D Knights, and R Blekhman. 2015. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Medicine 7:55. DOI:10.1186/s13073-015-1077-8).
This research strengthens our knowledge of the role played by the gut microbiome in cancer development and progression. It will help investigators trying to determine the causes and treatment of colorectal cancer, and may provide a basis for better diagnosis and treatment of the disease.
Timothy Starr (see the Starr Lab website) is an assistant professor in the Department of Obstetrics, Gynecology, and Women’s Health and is a member of the Masonic Cancer Center. He uses MSI resources to support his research into the genetic causes of cancer. Dan Knights (see the Knights Lab website) is an assistant professor in the Department of Computer Science and Engineering and is a member of the BioTechnology Institute. His work using MSI is helping to create a bioinformatics pipeline for linking genetic variation with bacteria. Ran Blekhman (see the Blekhman Lab website) is an assistant professor in the Departments of Genetics, Cell Biology, and Development and Ecology, Evolution, and Behavior. He is studying how cancer patient genomes, tumor genomes, and the microbiome interact to result in disease onset and evolution, and uses MSI for analyses of large genomic and metagenomic datasets. Lead author Dr. Michael Burns (post-doctoral associate) and co-author Joshua Lynch (graduate student) are both members of the Blekhman research group at MSI.
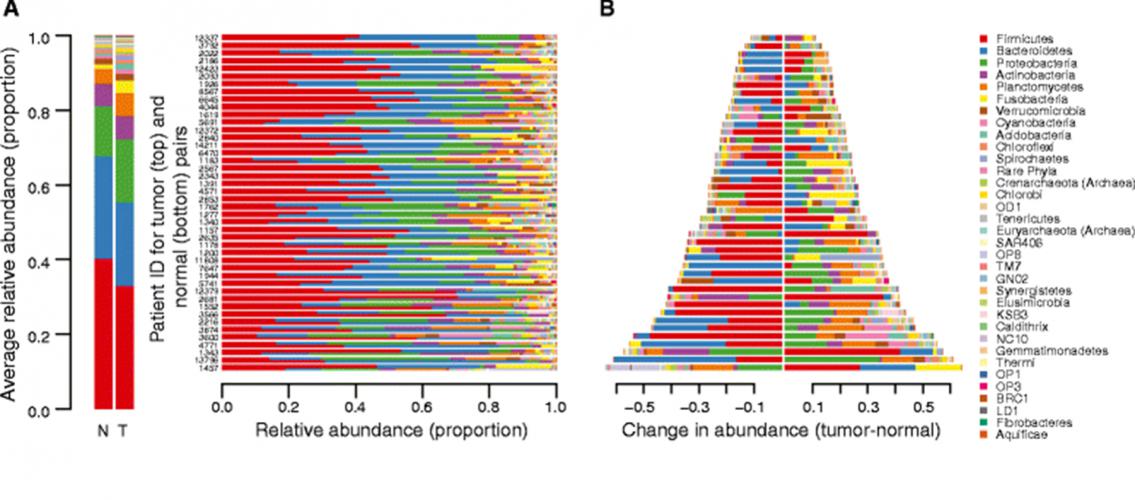
Image Description: Differences in bacterial and archaeal phyla within the normal and colorectal cancer microbiomes. A. Stacked bar plots indicating the proportional abundances of microbial phyla that are present at ≥1% in at least one sample. The averages across all normal samples (N) and tumor samples (T) are presented at the left. Rows are arranged as patient=matched tumor (top) and normal (bottom) pairs. Patient ID numbers are appended to the rights of the row pairs. B. The data in panel A presented as the difference in abundance (Tumor - Normal) for each phylum, sorted in the same patient order, where a value of 0 would indicate no difference. Note that the legend at the right is common to panels A and B. Image and description from Burns MB et al., Genome Medicine 2015 7:55 DOI:10.1186/s13073-015-1077-8.
posted on September 9, 2015